Bayesian Multimodel Inference
The abstract we submitted to the ESA Meeting was accepted a while back. Since we submitted it, Megan and I have been back in the field for some additional data collection and I've been doing some new analyses. Some of these new analyses are the result of my attendance at the Bayesian statistics workshop at US-IALE in Snowbird. Since then I've been learning more by picking the brain of a former advisor, George Perry, and doing a fair bit of reading (reading list with links at bottom). And of course, using my own data has helped a lot.
One of the main questions I'm facing, as many ecologists often do, is "which variables should be in my regression model?" This question lies at the core of model inference and assumes that it is appropriate to infer ecological process from data by searching for the single model that represents reality most accurately. However, as Link and Barker put it:
"It would be nice if there were no uncertainty about models. In such an ideal world, a single model would be available; the data analyst would be in the enviable position of having only to choose the best method for fitting model parameters based on the available data. The choice would be completely determined by the statistician's theory, a theory which regards the model as exact depiction of the process that generated the data.
"It is clearly wrong to use the data to choose a model and then to conduct subsequent inference as though the selected model were chosen a priori: to do so is to fail to acknowledge the uncertainties present in the model selection process, and to incestuously use the data for two purposes."
Thus, it usually more appropriate to undertake a process of multi-model inference and search for the 'best' possible model (given current data) rather than a single 'true' model. I've been looking into the use of Bayesian Model Averaging to address this issue. Bayesian approaches take prior knowledge (i.e., a probability distribution) and data about a system and combine them with a model to produce posterior knowledge (i.e., another probability distribution). This approach differs from the frequentist approach to statistics which calculates probabilities based on the idea of a (hypothetical) long-run of outcomes from a sequence of repeated experiments.
For example, estimating the parameters of a linear regression model using a Bayesian approach differs from a frequentist ordinary least squares (OLS) approach in two ways:
i) a Bayesian approach considers the parameter to be a random variable that might take a range of values each with a given probability, rather than being fixed with unknown probability,
ii) a Bayesian approach conditions the parameter estimate probability on the sample data at hand and not as the result of a set of multiple hypothetical independent samples (as the OLS approach does).
If there is little prior information available about the phenomena being modelled, 'uninformative priors' (e.g., a normal distribution with a relatively large variance about a mean of zero) can be used. In this case, the parameter estimates produced by the Bayesian linear regression will be very similar to those produced by regular OLS regression. The difference is in the error estimates and what they represent; a 95% confidence interval produced by a Bayesian analysis specifies that there is a 95% chance that the true value is within that interval given the data analyzed, whereas a 95% confidence interval from a frequentist (OLS) approach implies that if (hypothetical) data were sampled a large number of times, the parameter estimate for those samples would lie within that interval 95% of those times.
There has been debate recently in ecological circles about the merits of Bayesian versus frequentist approaches. Whilst some have strongly advocated the use of Bayesian approaches (e.g., McCarthy 2007), others have suggested a more pluralistic approach (e.g., Stephens et al. 2005). One of the main concerns with the approach of frequentist statistics is related to a broader criticism of the abuse and misuse of the P-value. For example, in linear regression models P-values are often used to examine the hypothesis that the slope of a regression line is not equal to zero (by rejecting the null hypothesis that is equal to zero). Because the slope of a regression line on a two-dimensional plot indicates the rate of change of one measure with respect to the other, a non-zero slope indicates that as one measure changes, so does the other. Consequently it is often inferred that a processes represented by one measure had an effect, or caused, the change in the other). However, as Ben Bolker points out in his excellent book:
"...common sense tells us that the null hypothesis must be false, because [the slope] can’t be exactly zero [due to the inherent variation and error in our data] — which makes the p value into a statement about whether we have enough data to detect a non-zero slope, rather than about whether the slope is actually different from zero."
This is not to say there's isn't a place for null hypothesis testing using P-values in the frequentist approach. As Stephens et al. argue, "marginalizing the use of null-hypothesis testing, ecologists risk rejecting a powerful, informative and well-established analytical tool." To the pragmatist, using whatever (statistical) tool available seems eminently more sensible than placing all one's eggs in one basket. The important point is to try to make sure that the hypotheses one tests with P-values are ecologically meaningful.
Back to Bayesian Model Averaging (BMA). BMA provides a method to account for uncertainty in model structure by calculating (approximate) posterior probabilities for each possible model (i.e., combination of variables) that could be constructed from a set of independent variables (see Adrian Raftery's webpage for details and examples of BMA implementation). The 'model set' is all possible combinations of variables (equal to 2n models, where n is the number of variables in the set). The important thing to remember with these probabilities is that it is the probability that the model is the best one from the model set considered - the probability of other models with variables not measured or included in the model set obviously can't be calculated.
The advantage over other model selection procedures like stepwise regression is that the output provides a measure of the performance of many models, rather than simply providing the single 'best' model. For example, here's a figure I derived from the output BMA provides:
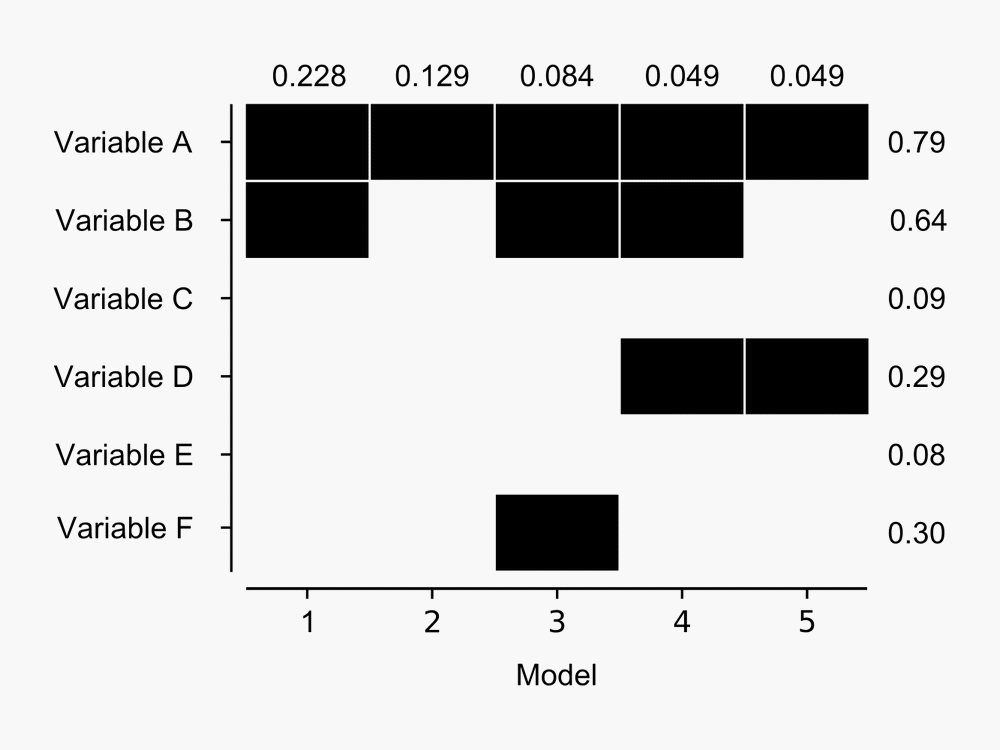
The figure shows BMA results for the five models with highest posterior probability of being the best candidate model from a hypothetical model set. The probability that each model is the best in the model set is shown at top for each model - Model 1 has almost 23% chance that it is the best model given the data available. Dark blocks indicate the corresponding variable (row) is included in a given model - so Model 1 contains variables A and B, whereas Model 2 contains Variable A only. Posterior probabilities of variables being included in the best model (in the model set) are shown to the right of the blocks - as we might expect given that Variable A is present in the five most probable models it has the highest chance of being included in the best model. Click for a larger image.
BMA also provides a posterior probability for each variable being included in the best candidate model. One of the cool things about the variable posterior probability is that it can be used to produce a weighted mean value from all the models for each variable parameter estimate, each with their own Bayesian confidence interval. The weight for each parameter estimate is the probability that variable is present in the 'best' model. Thus, the 'average' model accounts for uncertainty in variable selection in the best candidate model in the individual parameter estimates.
I've been using these approaches to investigate the potential factors influencing local winter white-tailed deer density in in managed forests of Michigan's Upper Peninsula. One of the most frequently used, and freely available, software packages for Bayesian statistics is WinBUGS. However, because I like to use R I've been exploring the packages available in that statistical language environment. Specifically, the BRugs package makes use of many OpenBUGS components (you actually provide R with a script in WinBUGS format to run) and the BMA package provides functionality for model averaging. We're in the final stages of writing a manuscript incorporating these analyses - once it's a bit more polished (and submitted) I'll provide an abstract.
Reading List
Model Inference: Burnham and Anderson 2002, Stephens et al. 2005, Link and Barker 2006, Stephens et al. 2007
Introduction to Bayesian Statistics: Bolker 2007 [webpage with chapter pre-prints and exercises here], McCarthy 2007
Discussion of BMA methods: Hoeting et al. 1999, Adrian Raftery's Webpage
Examples of BMA application: Wintle et al. 2003, Thomson et al. 2007
Criticisms of Stepwise Regression: James and McCulloch 1990, Whittingham et al. 2006
One of the main questions I'm facing, as many ecologists often do, is "which variables should be in my regression model?" This question lies at the core of model inference and assumes that it is appropriate to infer ecological process from data by searching for the single model that represents reality most accurately. However, as Link and Barker put it:
"It would be nice if there were no uncertainty about models. In such an ideal world, a single model would be available; the data analyst would be in the enviable position of having only to choose the best method for fitting model parameters based on the available data. The choice would be completely determined by the statistician's theory, a theory which regards the model as exact depiction of the process that generated the data.
"It is clearly wrong to use the data to choose a model and then to conduct subsequent inference as though the selected model were chosen a priori: to do so is to fail to acknowledge the uncertainties present in the model selection process, and to incestuously use the data for two purposes."
Thus, it usually more appropriate to undertake a process of multi-model inference and search for the 'best' possible model (given current data) rather than a single 'true' model. I've been looking into the use of Bayesian Model Averaging to address this issue. Bayesian approaches take prior knowledge (i.e., a probability distribution) and data about a system and combine them with a model to produce posterior knowledge (i.e., another probability distribution). This approach differs from the frequentist approach to statistics which calculates probabilities based on the idea of a (hypothetical) long-run of outcomes from a sequence of repeated experiments.
For example, estimating the parameters of a linear regression model using a Bayesian approach differs from a frequentist ordinary least squares (OLS) approach in two ways:
i) a Bayesian approach considers the parameter to be a random variable that might take a range of values each with a given probability, rather than being fixed with unknown probability,
ii) a Bayesian approach conditions the parameter estimate probability on the sample data at hand and not as the result of a set of multiple hypothetical independent samples (as the OLS approach does).
If there is little prior information available about the phenomena being modelled, 'uninformative priors' (e.g., a normal distribution with a relatively large variance about a mean of zero) can be used. In this case, the parameter estimates produced by the Bayesian linear regression will be very similar to those produced by regular OLS regression. The difference is in the error estimates and what they represent; a 95% confidence interval produced by a Bayesian analysis specifies that there is a 95% chance that the true value is within that interval given the data analyzed, whereas a 95% confidence interval from a frequentist (OLS) approach implies that if (hypothetical) data were sampled a large number of times, the parameter estimate for those samples would lie within that interval 95% of those times.
There has been debate recently in ecological circles about the merits of Bayesian versus frequentist approaches. Whilst some have strongly advocated the use of Bayesian approaches (e.g., McCarthy 2007), others have suggested a more pluralistic approach (e.g., Stephens et al. 2005). One of the main concerns with the approach of frequentist statistics is related to a broader criticism of the abuse and misuse of the P-value. For example, in linear regression models P-values are often used to examine the hypothesis that the slope of a regression line is not equal to zero (by rejecting the null hypothesis that is equal to zero). Because the slope of a regression line on a two-dimensional plot indicates the rate of change of one measure with respect to the other, a non-zero slope indicates that as one measure changes, so does the other. Consequently it is often inferred that a processes represented by one measure had an effect, or caused, the change in the other). However, as Ben Bolker points out in his excellent book:
"...common sense tells us that the null hypothesis must be false, because [the slope] can’t be exactly zero [due to the inherent variation and error in our data] — which makes the p value into a statement about whether we have enough data to detect a non-zero slope, rather than about whether the slope is actually different from zero."
This is not to say there's isn't a place for null hypothesis testing using P-values in the frequentist approach. As Stephens et al. argue, "marginalizing the use of null-hypothesis testing, ecologists risk rejecting a powerful, informative and well-established analytical tool." To the pragmatist, using whatever (statistical) tool available seems eminently more sensible than placing all one's eggs in one basket. The important point is to try to make sure that the hypotheses one tests with P-values are ecologically meaningful.
Back to Bayesian Model Averaging (BMA). BMA provides a method to account for uncertainty in model structure by calculating (approximate) posterior probabilities for each possible model (i.e., combination of variables) that could be constructed from a set of independent variables (see Adrian Raftery's webpage for details and examples of BMA implementation). The 'model set' is all possible combinations of variables (equal to 2n models, where n is the number of variables in the set). The important thing to remember with these probabilities is that it is the probability that the model is the best one from the model set considered - the probability of other models with variables not measured or included in the model set obviously can't be calculated.
The advantage over other model selection procedures like stepwise regression is that the output provides a measure of the performance of many models, rather than simply providing the single 'best' model. For example, here's a figure I derived from the output BMA provides:

The figure shows BMA results for the five models with highest posterior probability of being the best candidate model from a hypothetical model set. The probability that each model is the best in the model set is shown at top for each model - Model 1 has almost 23% chance that it is the best model given the data available. Dark blocks indicate the corresponding variable (row) is included in a given model - so Model 1 contains variables A and B, whereas Model 2 contains Variable A only. Posterior probabilities of variables being included in the best model (in the model set) are shown to the right of the blocks - as we might expect given that Variable A is present in the five most probable models it has the highest chance of being included in the best model. Click for a larger image.
BMA also provides a posterior probability for each variable being included in the best candidate model. One of the cool things about the variable posterior probability is that it can be used to produce a weighted mean value from all the models for each variable parameter estimate, each with their own Bayesian confidence interval. The weight for each parameter estimate is the probability that variable is present in the 'best' model. Thus, the 'average' model accounts for uncertainty in variable selection in the best candidate model in the individual parameter estimates.
I've been using these approaches to investigate the potential factors influencing local winter white-tailed deer density in in managed forests of Michigan's Upper Peninsula. One of the most frequently used, and freely available, software packages for Bayesian statistics is WinBUGS. However, because I like to use R I've been exploring the packages available in that statistical language environment. Specifically, the BRugs package makes use of many OpenBUGS components (you actually provide R with a script in WinBUGS format to run) and the BMA package provides functionality for model averaging. We're in the final stages of writing a manuscript incorporating these analyses - once it's a bit more polished (and submitted) I'll provide an abstract.
Reading List
Model Inference: Burnham and Anderson 2002, Stephens et al. 2005, Link and Barker 2006, Stephens et al. 2007
Introduction to Bayesian Statistics: Bolker 2007 [webpage with chapter pre-prints and exercises here], McCarthy 2007
Discussion of BMA methods: Hoeting et al. 1999, Adrian Raftery's Webpage
Examples of BMA application: Wintle et al. 2003, Thomson et al. 2007
Criticisms of Stepwise Regression: James and McCulloch 1990, Whittingham et al. 2006
Labels: Ecological, MichiganUP, Modelling, Statistical

This work by James D.A. Millington is licensed under a Creative Commons Attribution-Noncommercial-Share Alike 3.0 United States License.


0 Comments:
Post a Comment
<< Home